Motion Control
Thomas Burke Global Strategic Advisor for CC-Link Partner Association
Motion Control
How to Build Better Networkable DevicesHow to Build Better Networkable Devices
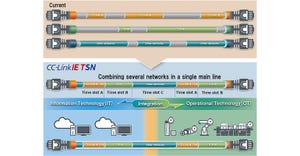
Industrial automation devices configured for time-sensitive networking may benefit from a single communications backbone for managing all Ethernet traffic.
Sign up for the Design News Daily newsletter.